Introducción a R y Rstudio
Profesores: Carlos M. González Alcón - Carlos Pérez González
(Dpto. Matemáticas, Estadística e Investigación Operativa - Universidad de La Laguna)
Librería RCommander y técnicas de análisis estadístico.
Nivel avanzado: Gráficos más complejos
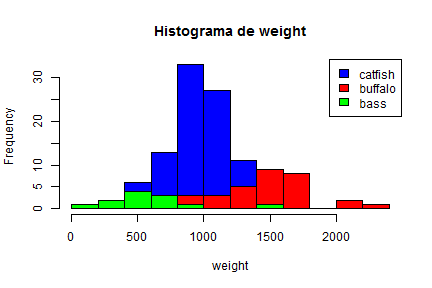
- Los gráficos histogramas también se pueden combinar
h1<-hist(ddt$weight, col="grey60",main="Histograma de weight",xlab="weight")
par(mfrow=c(1,1) )
with(ddt, hist(weight[species==1], breaks=h1$breaks, col="blue",main="Histograma de weight",xlab="weight" ))
with(ddt, hist(weight[species==2], breaks=h1$breaks, col="red",add=TRUE ))
with(ddt, hist(weight[species==3], breaks=h1$breaks, col="green",add=TRUE ))
legend( "topright", sort(levels(ddt$species_name),decreasing = TRUE) , fill=c("blue", "red","green") )
Otros gráficos en Rcmdr
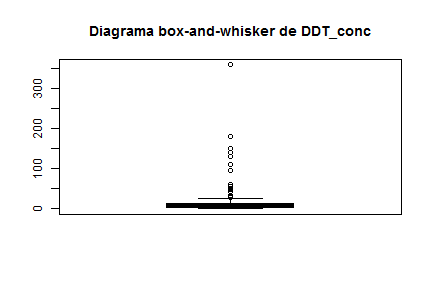
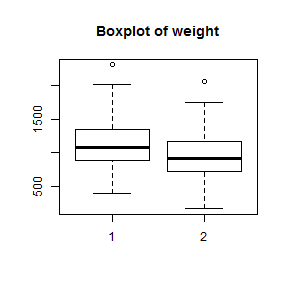
- Los gráficos de cajas y bigotes
boxplot( ddt$DDT_conc, main="Diagrama box-and-whisker de DDT_conc",col="gray")
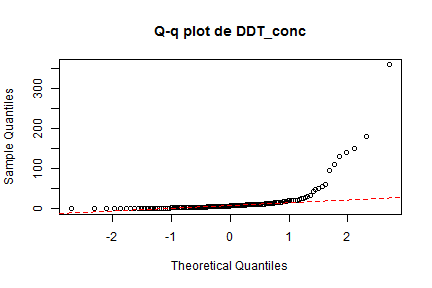
- Los gráficos de comparación de cuantiles (qq-plot)
qqnorm( ddt$DDT_conc, main="Q-q plot de DDT_conc")
qqline(ddt$DDT_conc, col="red", lty="dashed")
Ejercicio 3
Comparar el peso de los peces del grupo 1 con el peso de los del grupo 2.
- 1º PASO: ¿Se asume normalidad de la variable peso?
##
## Shapiro-Wilk normality test
##
## data: ddt$weight
## W = 0.9825, p-value = 0.06299
Si hay dudas en la normalidad, el t-test no se puede aplicar y hay que acudir a alternativas no paramétricas.
Ejercicio 4
Comparar el peso de los peces del grupo 1 con el peso de los del grupo 2.
- 2º PASO: ¿Se asume igualdad de varianzas de peso entre los grupos?
bartlett.test(weight ~ group, data=ddt)
##
## Bartlett test of homogeneity of variances
##
## data: weight by group
## Bartlett's K-squared = 0.13287, df = 1, p-value = 0.7155
Si las varianzas entre grupos son iguales, se debe especificar en opciones del t-test (aprox. Welch).
Ejercicio 5
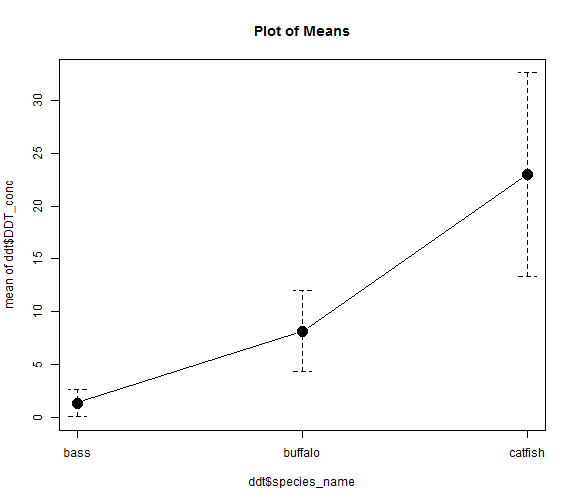
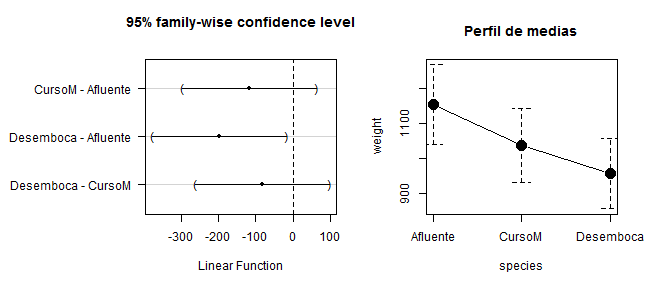
- EJEMPLO: Analizar las diferencias en el peso de los peces entre las diferentes localizaciones.
- 1º PASO: ¿Hay diferencias entre todos los niveles (especies)?
# Comprobamos si la varianza es constante
bartlett.test(weight ~ location_name, data=ddt)
##
## Bartlett test of homogeneity of variances
##
## data: weight by location_name
## Bartlett's K-squared = 0.78832, df = 2, p-value = 0.6742
# En caso de homoc. podemos hacer anova
anova.mod1 <- aov(weight ~ location_name, data=ddt)
summary(anova.mod1)
## Df Sum Sq Mean Sq F value Pr(>F)
## location_name 2 942700 471350 3.438 0.0349 *
## Residuals 141 19332838 137112
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
# Si no se asume var. cte. o no hay normalidad, recurrimos al test K.-W.
kruskal.test(weight ~ location_name, data=ddt)
##
## Kruskal-Wallis rank sum test
##
## data: weight by location_name
## Kruskal-Wallis chi-squared = 6.13, df = 2, p-value = 0.04665
Modelos estadísticos: Regresión
- La opción 'Ajuste de modelos' dispone de múltiples posibilidades para la creación de modelos estadísticos.
- Además, en la opción 'Modelos' se pueden llevar a cabo muchos análisis habituales en el estudio de los mismos.
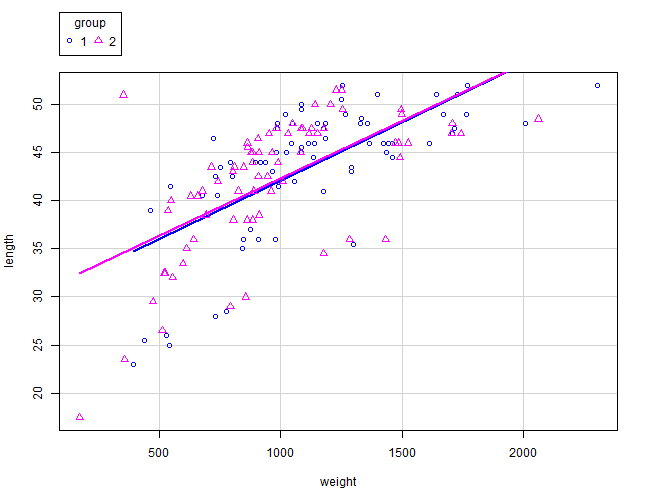
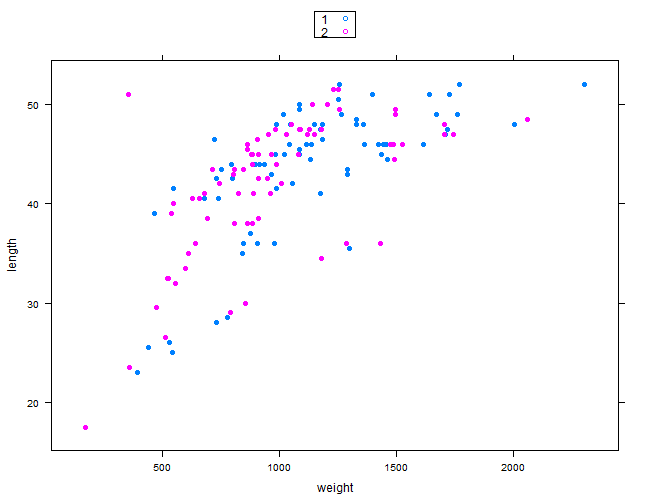
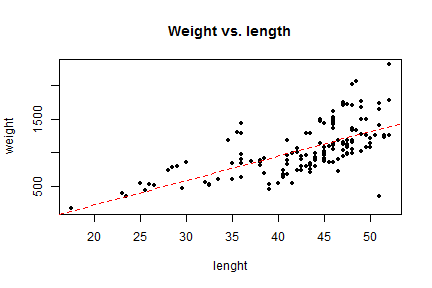
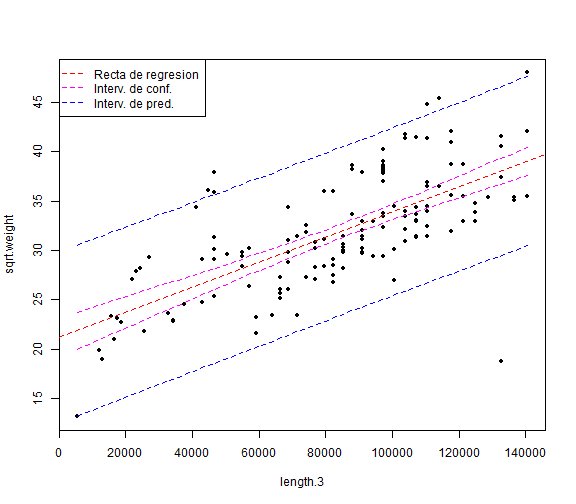
- En particular, veamos como determinar un modelo de regresión para estudiar la relación entre la longitud y el peso de los peces.
regmod.1 <- lm(weight~length, data=ddt)
summary(regmod.1)
##
## Call:
## lm(formula = weight ~ length, data = ddt)
##
## Residuals:
## Min 1Q Median 3Q Max
## -989.96 -189.45 -49.51 193.68 923.22
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -483.672 150.497 -3.214 0.00162 **
## length 35.816 3.471 10.319 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 285.7 on 142 degrees of freedom
## Multiple R-squared: 0.4285, Adjusted R-squared: 0.4245
## F-statistic: 106.5 on 1 and 142 DF, p-value: < 2.2e-16
Análisis de modelos
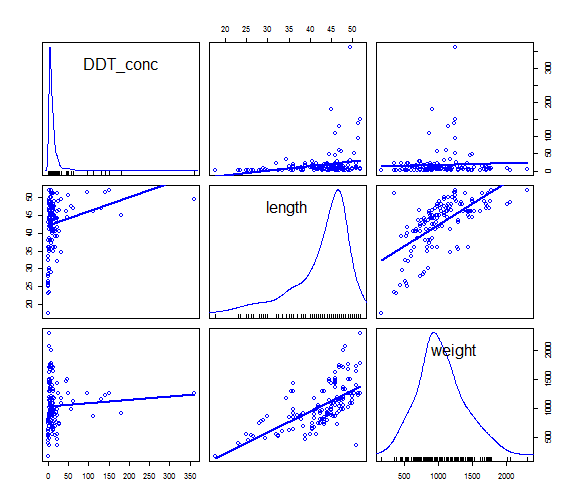
- Podemos obtener la correlación de las variables de interés en 'Estadísticos->Resúmenes'.
cor(ddt[,c("length","weight")], use="complete")
## length weight
## length 1.0000000 0.6546113
## weight 0.6546113 1.0000000
- La representación gráfica de un modelo simple es posible en 'Gráficas->Diagrama de dispersión' seleccionando la opción de mínimos cuadrados. Pero se puede hacer también con
plot() y abline()
plot(ddt$length, ddt$weight, main="Weight vs. length", xlab="lenght", ylab="weight",
pch=20)
abline(regmod.1,col="red",lty="dashed")
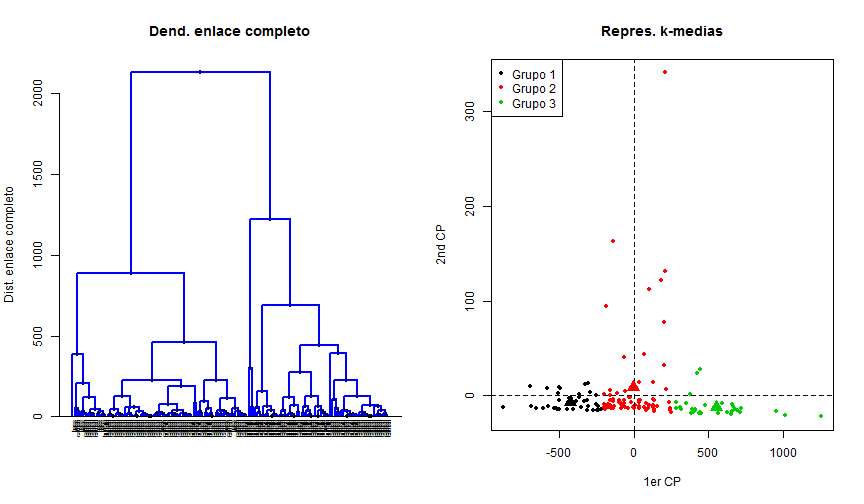
Modelos estadísticos: Clustering
- La opción 'Análisis dimensional->Análisis de agrupación' permite aplicar técnicas de cluster.
require(devtools)
source_gist("a4b2ed204d01bc12d952",filename="plotCluster.R")
par(mfrow=c(1,2))
par(mfrow=c(1,2))
plotCluster(1,ddt[,c("length","weight","DDT_conc")],ddt$species_name)
plotCluster(2,ddt[,c("length","weight","DDT_conc")],ddt$species_name)